RESEARCH
Transposon Group
For Research
Recently, our lab is interested in investigating transposable elements or "jumping genes" that comprise ~45% of the human genome. These elements can mobilize within the genome resulting in intra- and inter-genetic variation, and cause gene disruption associated with sporadic genetic diseases. Specifically, we are interested in uncovering the mechanisms that regulate LINE-1 retrotransposon.
Studies of a human retrotransposon L1
What is L1?
Transposable elements (often called ‘jumping genes’) mobilize to other genomic loci and comprise ~45% of our genome. Human Long INterspersed Element-1 (LINE-1 or L1) retrotransposons comprise ~17% of the human genome and are known as the only autonomous retrotransposon in the human genome. Although the majority of L1s cannot retrotranspose due to 5’ truncation or other mutations, ~80-100 copies of L1s are expected to be highly active in the human genome. L1 retrotransposition occurs in various types of cells including germ and somatic cells, indicating that L1 generates inter- and intra-genetic variation. L1 can also mediate retrotransposition of non-autonomous elements such as Alu and SVA elements (SINEs) in trans. In other words, about one third of our genome is derived from L1-mediated retrotransposition.
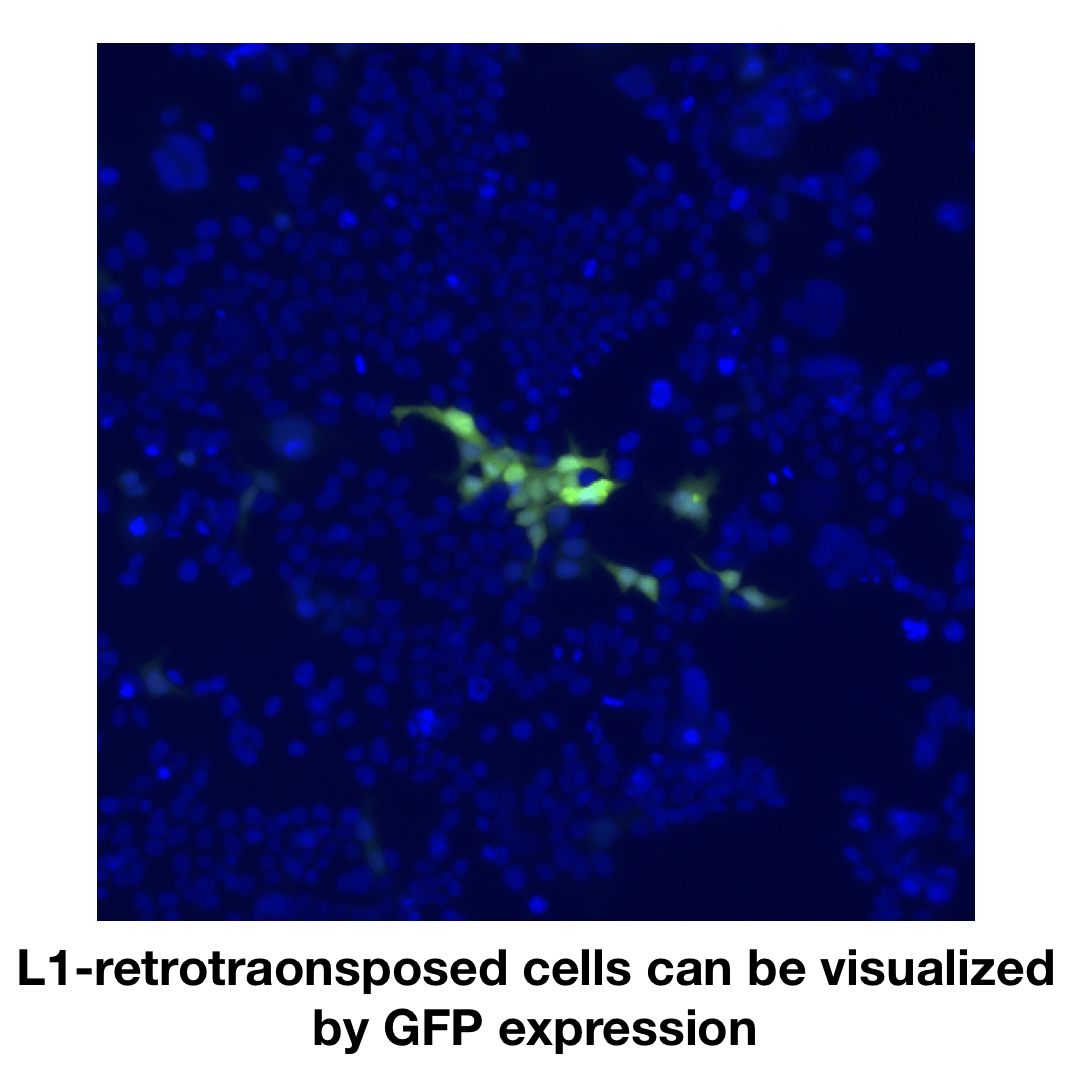
It had long been a mystery whether L1 was actually able to move in the human genome, but finally it was discovered that L1 causes disease-associated gene disruption in patients with Hemophilia A and colon cancer (Kazazian et al., Nature 1988; Miki et al., Cancer Res. 1992). In 1996, L1 mobilization was recapitulated using an engineered L1 construct in cultured human cells and termed the L1 retrotransposition assay (Moran et al., Cell 1996; Feng et al., Cell 1996). This breakthrough technique allows the investigation of the molecular mechanisms of L1 retrotransposition. Using this technique, it has been reported that L1 causes not only gene disruption but also genomic rearrangement associated with large deletion, inversion or perhaps translocation (Gilbert et al., Cell 2002; Symer et al., Cell 2002).
What cellular factors regulate LINE-1 retrotransposition?
L1 encodes two open reading frames (ORF1 and ORF2). ORF1p is an RNA binding protein with nucleic chaperon activity. ORF2p is the L1 catalytic component that possesses endonuclease (EN) and reverse transcriptase (RT) activities. After transcription, ORF1p and ORF2p are translated and bind their own RNA. L1s retrotranspose by a copy and paste mechanism called target-site primed reverse transcription (TPRT). ORF2p EN introduces a nick on the genomic DNA, liberating a 3’ hydroxyl group, which is necessary to initiate reverse transcription by ORF2p RT activity. However, the molecular mechanism of L1 integration during and after the TPRT reaction is largely unknown; what cellular factors help and repress L1 integration (DNA processing, fill-in and ligation)? What DNA damage response senses the damage created by L1? Where can L1s jump to in our genome? Thus, much remains to be solved concerning how L1s mobilize in the human genome.
Retrotransposons should have co-evolved with host cellular factors that suppress their mobilization associated with genomic instability. This is often referred to as an “evolutionary arms race”. In general, such host factors are also involved in other cellular activities, such as anti-viral defense responses, epigenetic regulation and possibly DNA repair. Therefore, the more we know about the molecular mechanisms of L1 retrotransposition, the better we can understand host defense mechanisms against both environmental and intrinsic genome stresses including virus infection and DNA damage. Furthermore, our study could contribute to developing a new method to screen chemical inhibitors that can minimize the threat of L1s to genome integrity. We are currently investigating what cellular factors regulate L1 retrotransposition molecularly by using the L1 retrotransposition assay and mass spectrometry analysis of L1-interacting proteins.